Advancing Epigenetics Research with ChIP-Exo-Seq Validated Antibodies
Author: Laura Pozzi PhD, Scientific Content Manager, Atlas Antibodies AB, Sweden
What is epigenetic research?
Epigenetic research is the study of heritable changes in gene function that do not involve changes to the underlying DNA sequence. In other words, epigenetics explores how gene activity is regulated without altering the actual genetic code. These changes can turn genes "on" or "off", affecting how cells read genes and produce proteins.
Epigenetic research aims to understand how cells with the same DNA can have very different identities and functions, and how gene expression patterns are passed on through cell divisions or altered by external influences. It’s a powerful lens through which we study gene regulation, disease mechanisms, inheritance, and potential therapies.
Why epigenetics matters in research?
Epigenetic regulation is dynamic, reversible, and influenced by developmental signals, environment, diet, stress, and even toxins. This makes it highly relevant in:
- Developmental biology – understanding how cells differentiate into specific types.
- Cancer research – tumors often show abnormal methylation or histone marks.
- Neuroscience – epigenetic changes impact learning, memory, and mental health.
- Aging and longevity – epigenetic clocks measure biological age based on DNA methylation patterns.
- Pharmacology – epigenetic drugs (like HDAC inhibitors or DNMT inhibitors) are being developed to treat diseases.
Understanding how genes are regulated at the chromatin level is essential for epigenetics research.
Chromatin Immunoprecipitation (ChIP) is a powerful technique that enables researchers to study interactions between DNA and proteins allowing the precisely mapping of transcription factor binding sites, histone modifications, and chromatin accessibility, key factors in understanding gene regulation and epigenetic mechanisms.
However, traditional ChIP approaches often struggle with background noise and resolution limitations. ChIP-Exo-Seq, an advanced version of ChIP, overcomes these challenges by integrating exonuclease digestion to provide single-nucleotide resolution mapping of DNA-protein interactions. ChIP-exo introduces an exonuclease digestion step after immunoprecipitation, trimming DNA right up to the protein-DNA crosslink point. This allows base-pair resolution mapping of binding sites.
For ChIP techniques, the choice of antibodies is critical. Non-specific or poorly characterized antibodies can lead to misleading results and poor reproducibility. Using validated antibodies ensures specificity, sensitivity, and consistent performance, factors that are essential for high-confidence chromatin research.
ChIP vs. ChIP-Exo-Seq overview
-
Traditional ChIP: A Powerful but Broad Approach
ChIP has been a cornerstone technique in epigenetics, allowing researchers to identify protein-DNA interactions and histone modifications. The process involves cross-linking proteins to DNA, fragmenting chromatin, and using antibodies to immunoprecipitate target protein-DNA complexes. These complexes are then analyzed via qPCR (ChIP-qPCR) for specific regions or next-generation sequencing (ChIP-Seq) for genome-wide profiling.
While ChIP-Seq has revolutionized epigenetics research, it still has resolution limitations and background noise. Since DNA fragmentation is random, the exact protein-binding site can be difficult to determine with precision. Additionally, non-specific antibody binding can generate false positives, making antibody validation a key factor in data reliability.
-
ChIP-Exo-Seq: Precision in Chromatin Analysis
ChIP-Exo-Seq is an improved version of ChIP-Seq that integrates exonuclease digestion (exo) to enhance resolution and specificity. After immunoprecipitation, an exonuclease trims unprotected DNA, leaving behind only the DNA fragments directly bound to proteins. This process allows researchers to pinpoint the precise nucleotide sequence where a protein interacts with DNA, eliminating background noise and increasing confidence in binding site identification.
-
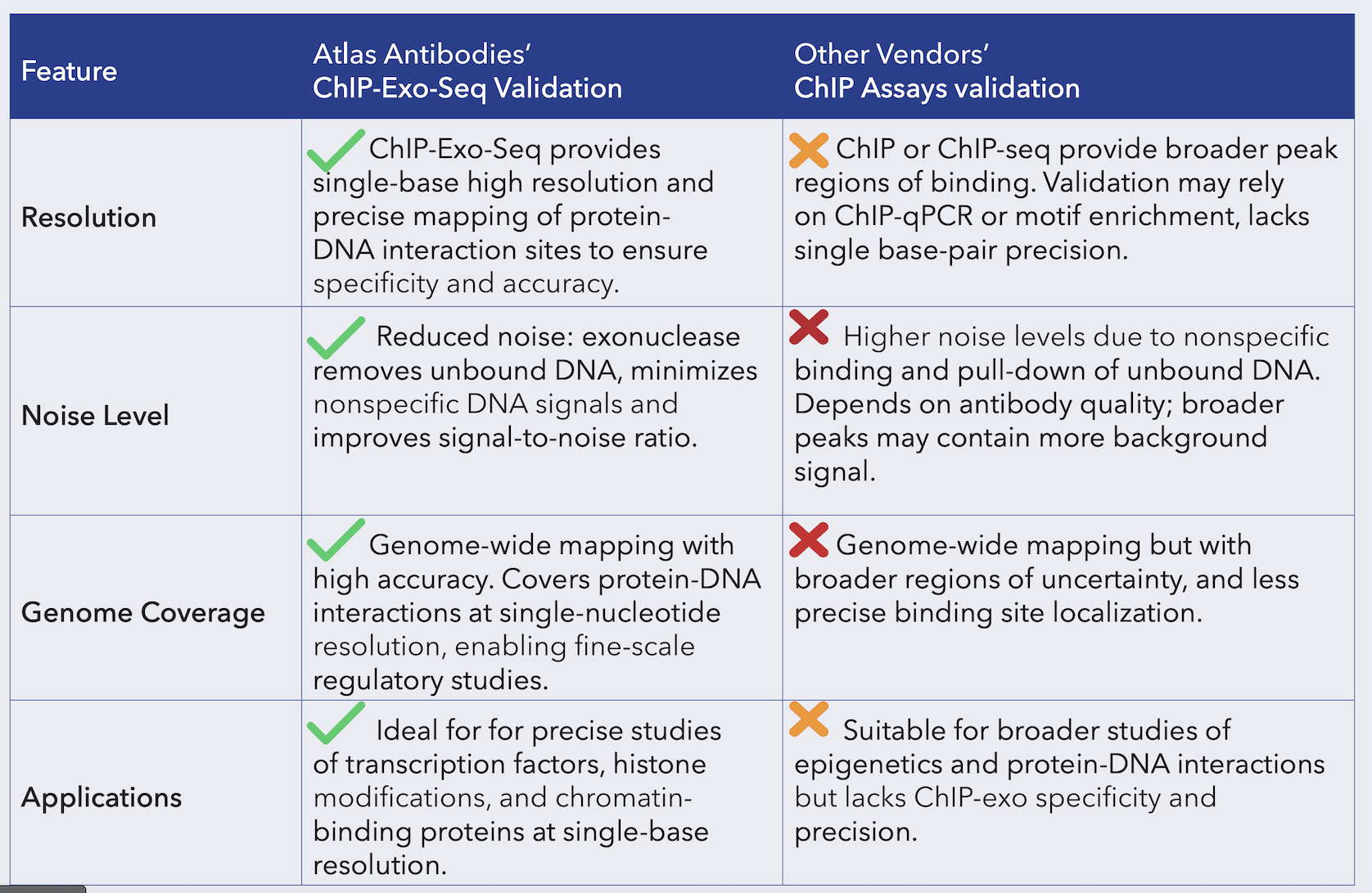
Compared to traditional ChIP, ChIP-Exo-Seq offers several advantages including:
· Higher resolution – Achieves near base-pair accuracy.
· Reduced background – Eliminates excess DNA, improving signal clarity.
· More accurate transcription factor binding site detection (crucial for understanding gene regulation).
By adopting ChIP-Exo-Seq with validated antibodies, researchers can gain deeper insights into chromatin dynamics, regulatory elements, and epigenetic modifications with unprecedented accuracy.
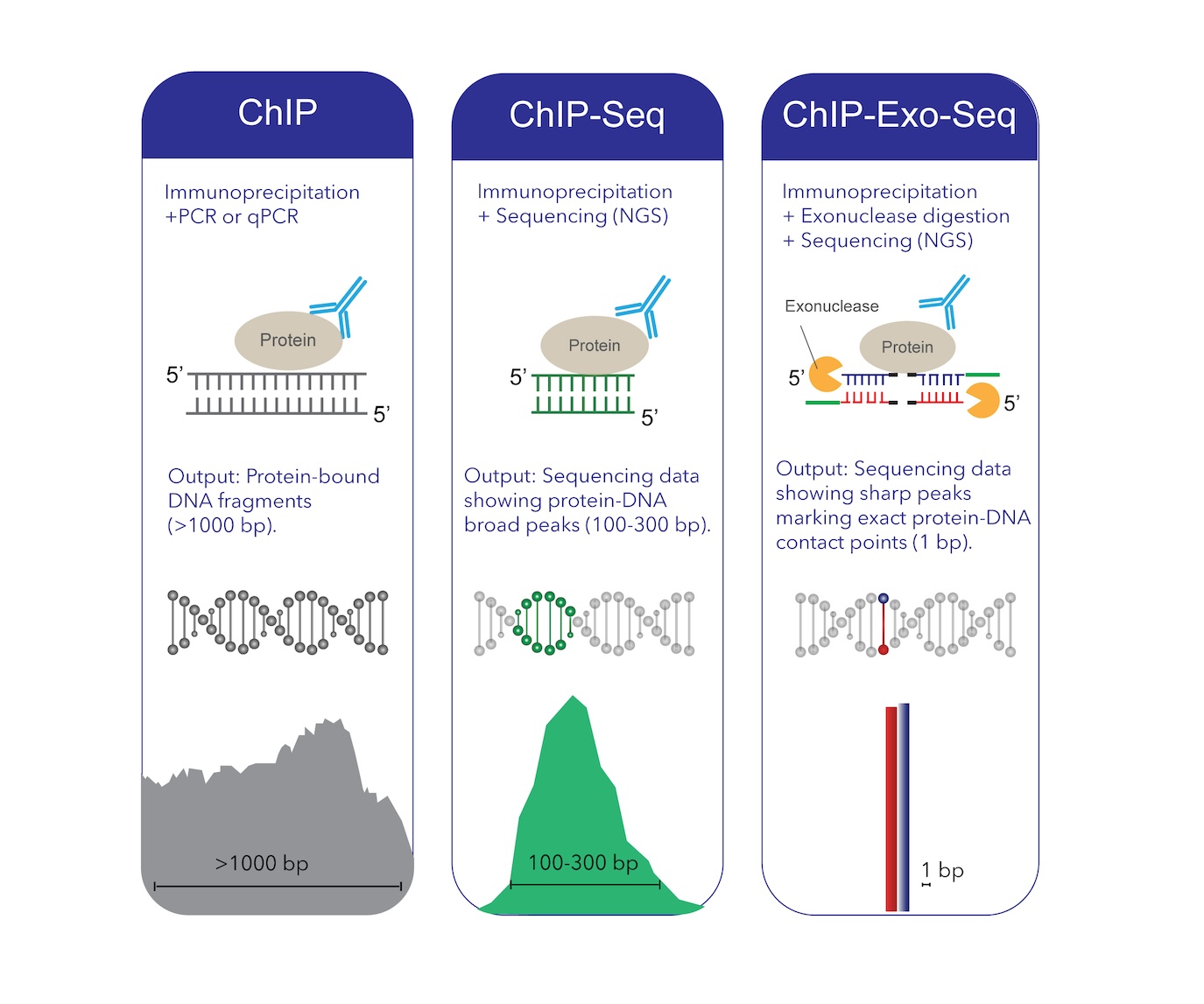
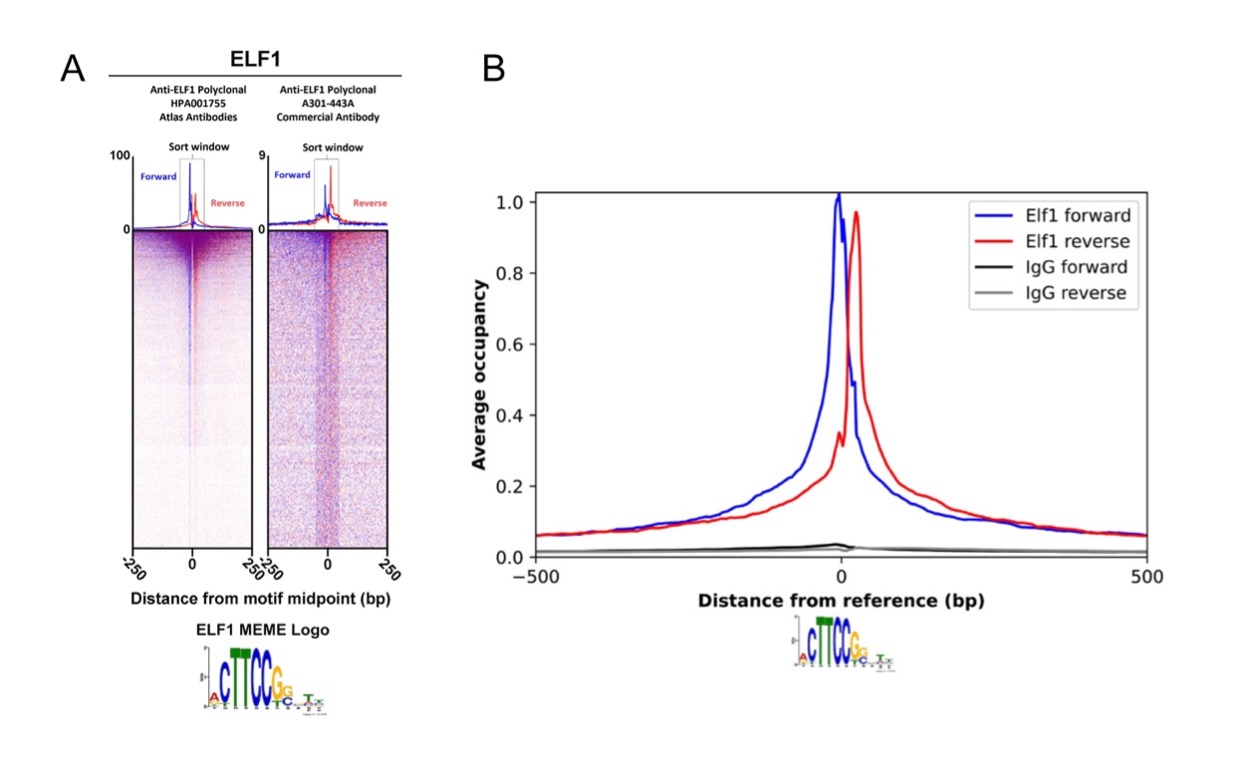
Figure 1 - Unmatched Precision ChIP-Exo-Seq combines Chromatin Immunoprecipitation (ChIP) with exonuclease digestion (exo) and high-throughput NGS sequencing (seq), offering single-base resolution for more accurate and specific mapping of protein-DNA binding sites compared to traditional ChIP or ChIP-Seq. It's ideal for studying protein-DNA interactions, epigenetics, and transcriptional regulation.
Applications of ChIP-Exo-Seq in epigenetics research
Mapping Transcription Factor Binding with High Precision: Transcription factors (TFs) play a critical role in gene expression regulation by binding specific DNA sequences. Traditional ChIP-Seq provides broad binding regions, but ChIP-Exo-Seq allows single-nucleotide precision, enabling researchers to accurately determine where TFs bind and how they influence transcription. This level of detail is essential for understanding gene regulatory networks in normal development and disease.
Studying Histone Modifications and Chromatin Dynamics: Histone modifications dictate chromatin structure and gene accessibility. ChIP-Exo-Seq enables high-resolution mapping of histone marks, distinguishing between active, poised, and repressed chromatin states. This is particularly valuable in studies of cell differentiation, cancer epigenetics, and developmental biology.
Epigenetic Biomarker Discovery and Drug Development: Epigenetic modifications are increasingly recognized as biomarkers for disease. ChIP-Exo helps identify disease-associated regulatory elements, aiding in the development of targeted therapies and epigenetic drugs. By mapping epigenetic changes at high resolution, researchers can pinpoint novel drug targets and biomarkers for conditions like cancer, neurological disorders, and autoimmune diseases.